Functional genetic screens for enhancer elements. (Nature Biotechnology 2015)
Enhancers are genomic domains that regulate transcription of distantly located genes through chromatin looping. They function as binding platforms for transcription factors and are characterized by specific chromatin signatures of histone methylation and acetylation. Only a small subset of all enhancers is active at a given space and time during development, indicating a tight regulation of enhancer activity to control gene expression. Intriguingly, recent studies have indicated that single nucleotide polymorphisms, large-scale genomic rearrangements and somatic mutations can affect enhancer activity and contribute to tumorigenesis. Moreover, the relevance of enhancers in cancer biology is highlighted by the fact that transcription factors and other enhancer-associated factors are frequently mutated in tumors, and targeting these factors by small molecule inhibitors yields great therapeutic potential.
To date, however, systematic identification of enhancer functions is hampered by the lack of tools to perform unbiased functional genetic screens. We therefore devised a novel approach for this purpose by utilizing the recently developed genome editing CRISPR-Cas9 tool. We present two distinct genetic screens to identify and characterize functional enhancers in their native environment. As a result, we identified several functional enhancer elements that are required for either inhibition or stimulation of proliferation by the tumor suppressor p53- and the mitogen ERα, respectively. Finally, we show that a genomic CRISPR-Cas9 tiling screen can precisely map functional domains within enhancer elements. Altogether, our results allow for the first time to expand the utility of CRISPR-Cas9 to explore the functions of the non-coding genome under normal and pathological conditions. .
To date, however, systematic identification of enhancer functions is hampered by the lack of tools to perform unbiased functional genetic screens. We therefore devised a novel approach for this purpose by utilizing the recently developed genome editing CRISPR-Cas9 tool. We present two distinct genetic screens to identify and characterize functional enhancers in their native environment. As a result, we identified several functional enhancer elements that are required for either inhibition or stimulation of proliferation by the tumor suppressor p53- and the mitogen ERα, respectively. Finally, we show that a genomic CRISPR-Cas9 tiling screen can precisely map functional domains within enhancer elements. Altogether, our results allow for the first time to expand the utility of CRISPR-Cas9 to explore the functions of the non-coding genome under normal and pathological conditions. .
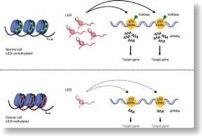
eRNAs are required for p53-dependent enhancer activity and gene transcription. (Molecular Cell 2013)
Binding within or nearby target genes involved in cell proliferation and survival enables the p53 tumor suppressor gene to regulate their transcription and cell-cycle progression. Using genome-wide chromatin-binding profiles, we describe binding of p53 also to regions located distantly from any known p53 target gene. Interestingly, many of these regions possess conserved p53-binding sites and all known hallmarks of enhancer regions. We demonstrate that these p53-bound enhancer regions (p53BERs) indeed contain enhancer activity and interact intrachromosomally with multiple neighboring genes to convey long-distance p53-dependent transcription regulation. Furthermore, p53BERs produce, in a p53-dependent manner, enhancer RNAs (eRNAs) that are required for efficient transcriptional enhancement of interacting target genes and induction of a p53-dependent cell-cycle arrest. Thus, our results ascribe transcription enhancement activity to p53 with the capacity to regulate multiple genes from a single genomic binding site. Moreover, eRNA production from p53BERs is required for efficient p53 transcription enhancement.
.