The tumor-suppressive functions of the human INK4A locus
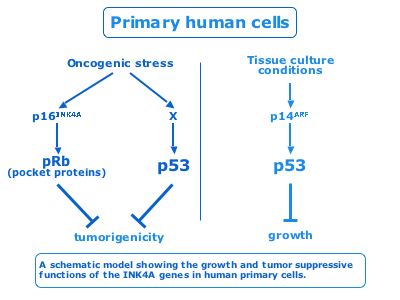
Tumor suppressors are genes found inactivated by mutations or deletions in a subset of human cancers. However, each individual tumor contains many genetic alterations, complicating the study of the contribution of each variation to tumorigenesis. We studied the tumor-suppressive functions of the human INK4A locus, which encodes for the p16INK4A and p14ARF genes and is mutated in many cancers. In mouse model systems, p19ARF plays an important protective role from oncogene induced transformation whereas p16INK4A loss causes only limited induction of tumors. Using pSUPER technology we showed that p16INK4A, but not p14ARF, is the major tumor suppressor of the human INK4A locus in response to oncogenic stress. We are currently engaged in the elucidation of the p14ARF independent pathways that are activated by oncogenic stress.
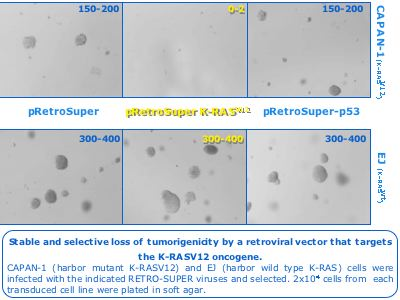
The pSUPER system is sensitive to mutations to the extent that a single nucleotide mismatch in the targeting sequence abrogates its ability to suppress gene expression. This feature opened opportunities for developing new therapy approaches by targeting disease-derived transcripts (e.g oncogenes) with dominant activating mutations. One such example is the occurrence of mutated RAS alleles in many human tumors. To examine the role of RAS mutants in tumor maintenance we have developed a pSUPER tool to specifically target oncogenic K-RASV12 allele without affecting its wild type counterpart. We showed that this tool powerfully inhibits the tumorigenicity of cancer cell lines that harbor this exact type of genetic alteration. As K-RASV12 is a frequent event in human cancer, the pSUPER-K-RASV12 and similar expression vectors can be used now to identify essential genetic events in human cancers and also may possibly serve as genetic tools for cancer therapy.
Identification and characterization of novel tumor suppressor genes
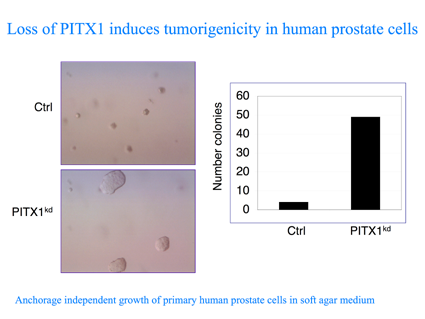
In a complementary approach, we are using the pSUPER RNAi library to target putative tumor suppressor in order to identify and define combinations of genetic events that are capable of converting primary human cells to cancerous cells. To uncover novel tumour-suppressor genes affecting the RAS pathway, we screened an RNA-interference library for knockdown constructs that transform human primary cells in the absence of oncogenic RAS. We report the identification of PITX1, whose inhibition induces the RAS pathway and tumorgenicity. Interestingly, we observed low expression of PITX1 in prostate and bladder tumours and in colon cancer cell lines containing wild-type RAS. Restoration of PITX1 in the colon cancer cells inhibited tumorigenicity, in a wild-type RAS dependent manner. Finally, we identified the mechanism through which PITX1 affect RAS activity. PITX1 activates RASAL1, a member of the RAS-GTPase-activating protein family, by direct binding to its promoter. Altogether, our results outline an approach to uncover tumour suppressor genes that function as inhibitors of the RAS pathway, which can also be used for other cancer causing pathways, such as the p53 and pRb.
The data extracted from such study enables us to understand the function of various tumor suppressors in tumor progression and to identify new critical genes required for cancer development in humans.