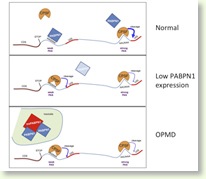
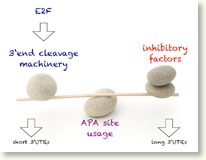
Regulation of alternative cleavage and polyadenylation of mRNAs. (Cell 2012)
3’end of most message RNAs (mRNAs) is cleaved and polyadenylated, a process that is required for mRNA function. Recent discoveries revealed that a large proportion of human genes contain more than one polyadenylation site. Therefore, alternative cleavage and polyadenylation (APA) is a widespread phenomenon that generates mRNAs with alternative 3’ends. Potentially, APA generates isoforms differing in their coding and non-coding regions, thereby affecting gene function and its regulation by miRNAs and RBPs. Thus, APA provides an important regulatory layer of gene expression. Interestingly, very strong genome-wide switches in APA are observed when cells are stimulated to proliferate, differentiate, and during cancer progression. However, how APA is regulated and what is its function, are largely unknown.
To identify regulators of APA we developed a reporter-based RNAi screen and devised a genome-wide approach to detect 3’ends of mRNAs (named 3’Seq). With these tools we identified the gene PABPN1 as a regulator of APA. Loss-of PABPN1 resulted in extensive 3'UTR shortening and a compromised miRNA-mediated repression. Interestingly mutations in PABPN1 causes the autosomal dominant oculopharyngeal muscular dystrophy (OPMD). Intriguingly, the expression of mutant PABPN1 in both a mouse model of OPMD and human cells elicited 3’UTR shortening, linking for the first time APA with a genetic disease. We conclude that PABPN1 is a suppressor of APA (Top Figure).
Additionally, we explored, on a transcriptome-wide scale, APA events that are associated with cancer. First, we comprehensively mapped APA events associated with cellular proliferation and transformation, and cancer. Second, we demonstrated that E2F-mediated transcriptional regulation of 3’-end processing genes is one of the key mechanisms that links APA to proliferation. Increase in E2F activity enhances 3’-end-processing gene expression that results in more potent usage of alternative polyadenylation sites (Bottom Figure).
.